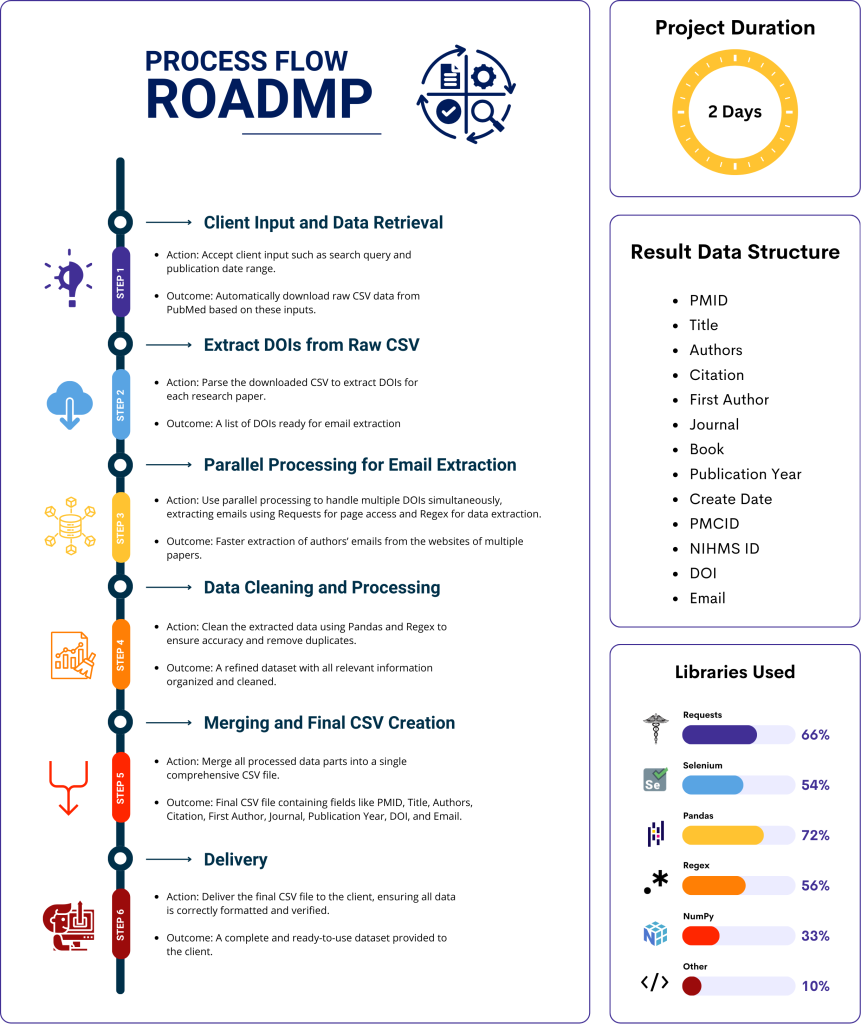
In this project, I developed a Python program to automate the extraction of author emails and additional metadata from scientific papers on PubMed. The program takes user inputs such as search queries and publication date range, downloads raw data in CSV format, and processes each paper’s website using DOI links to extract relevant information. By employing parallel processing, the program optimizes performance, handling large datasets efficiently and outputting cleaned, structured data in a new CSV file. This tool significantly accelerates the process of data extraction, making it a valuable asset for researchers and professionals needing bulk data from PubMed.
PubMed Interface Overview
This section showcases the final result of the automated process, where news articles scraped from multiple sources are published on a WordPress website. Each post is enhanced for SEO using GPT models to optimize the content, meta description, and focus keywords. The screenshots display the homepage featuring the latest news updates and the detailed post page, reflecting how the data is presented to users.
Project Information
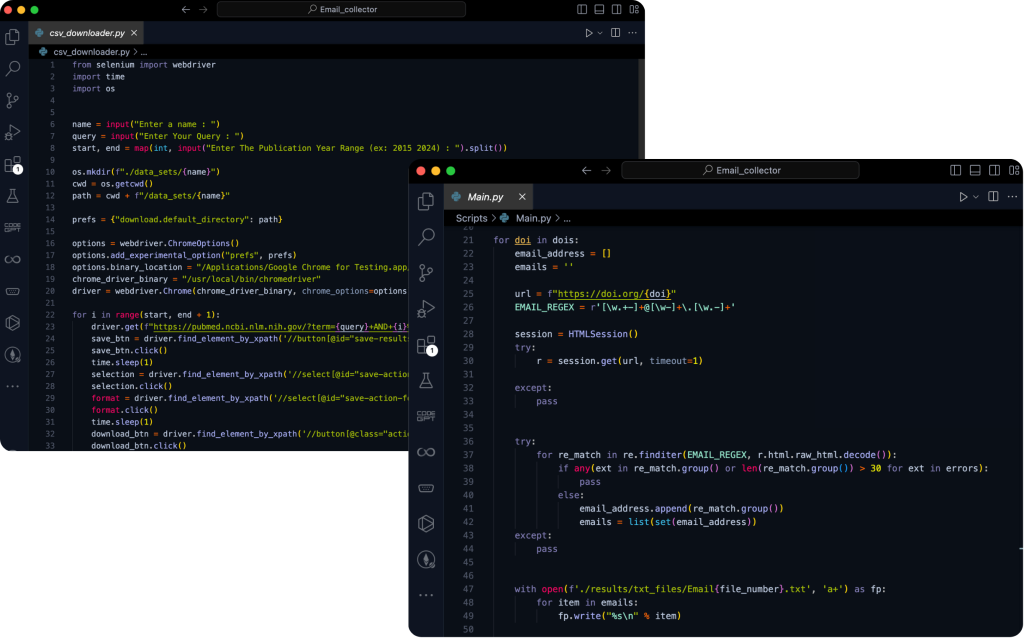
Code Implementation
Below is a snapshot of the core code used in this project. This segment highlights the extraction process, including fetching DOI links, parsing HTML content, and applying regex for email identification. The implementation showcases the use of Python libraries like Requests, Selenium, Pandas, and NumPy, integrated into a seamless workflow that handles both data extraction and error management efficiently.
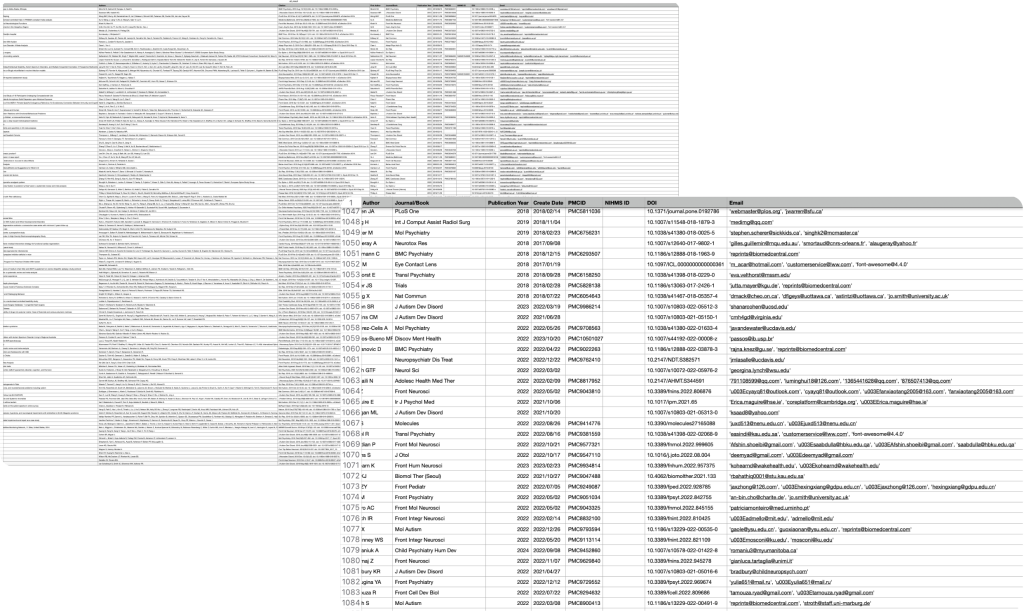
Extraction Results
Here is an example of the result data generated by the program. The output is a well-organized CSV file containing crucial information such as author emails, publication details, and identifiers. This data format allows for easy analysis and integration into further research workflows, providing immediate value to users requiring structured information from PubMed.